Seed the initial infections, and other compartments, in a spatial simulation
Source:R/SVEIRD.BayesianDataAssimilation.R
castSeedDataQueensNeighbourhood.RdThe Moore neighbourhood around the locations given in the seed data is affected with the initial values recorded in the provided seed data. The infected and exposed compartments are cast in a Moore neighbourhood of cells about the geographical coordinates recorded locations, while other compartments are cast directly into the cell corresponding exactly to the location, not a Moore neighbourhood.
Arguments
- layers
The SpatRaster object with layers Susceptible, Vaccinated, Exposed, Infected, Recovered, and Dead.
- seedData
a dataframe like the following example; the compartment columns are the initial values.
Location Latitude Longitude Vaccinated Exposed Infected Recovered Dead Beni 0.49113 29.47306 0 24 12 0 4 Butembo 0.140692 29.335014 0 0 0 0 0 Mabalako 0.461257 29.210687 0 0 0 0 0 Mandima 1.35551 29.08173 0 0 0 0 0- neighbourhood.order
The order of the queen's neighbourhood over which to distribute the Exposed and Infected intial state variable; zero is no neighbourhood, and corresponds to only the same cell where the queen already is. Higher orders follow the simple formula
(2 * x + 1)^2to determine the number of cells over which to evenly disperse the exposed and infected state variables. All other state variables are placed directly in the grid cell corresponding to the health zone.
Examples
subregionsSpatVector <- terra::vect(
system.file(
"extdata",
## COD: Nord-Kivu and Ituri (Democratic Republic of Congo)
"subregionsSpatVector",
package = "spatialEpisim.foundation",
mustWork = TRUE
)
)
susceptibleSpatRaster <- terra::rast(
system.file(
"extdata",
"susceptibleSpatRaster.tif", # Congo population
package = "spatialEpisim.foundation",
mustWork = TRUE
)
)
layers <- getSVEIRD.SpatRaster(subregionsSpatVector,
susceptibleSpatRaster,
aggregationFactor = 10)
data(initialInfections.fourCities, package = "spatialEpisim.foundation")
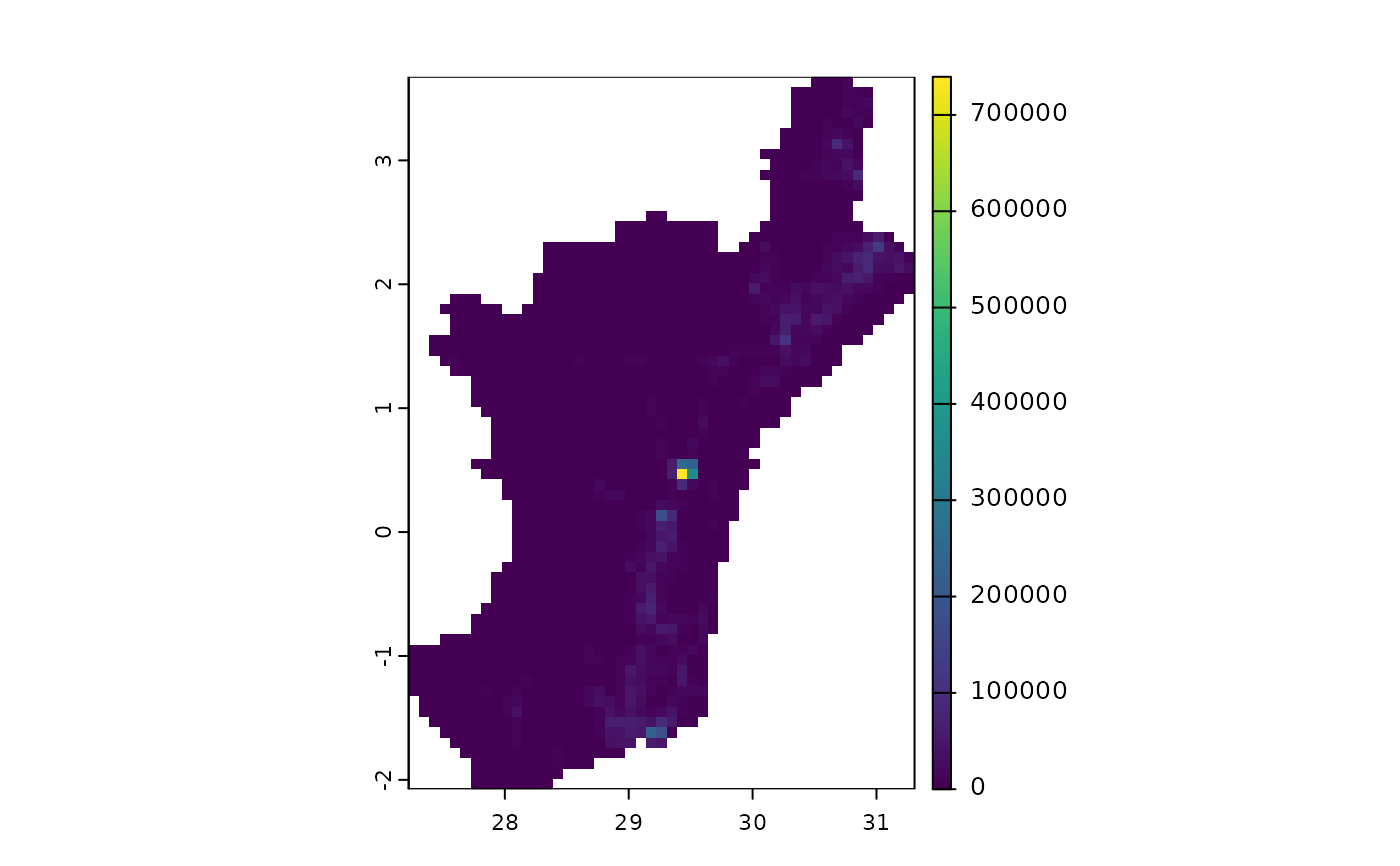
terra::plot(castSeedDataQueensNeighbourhood(layers, initialInfections.fourCities, 0)$Susceptible)
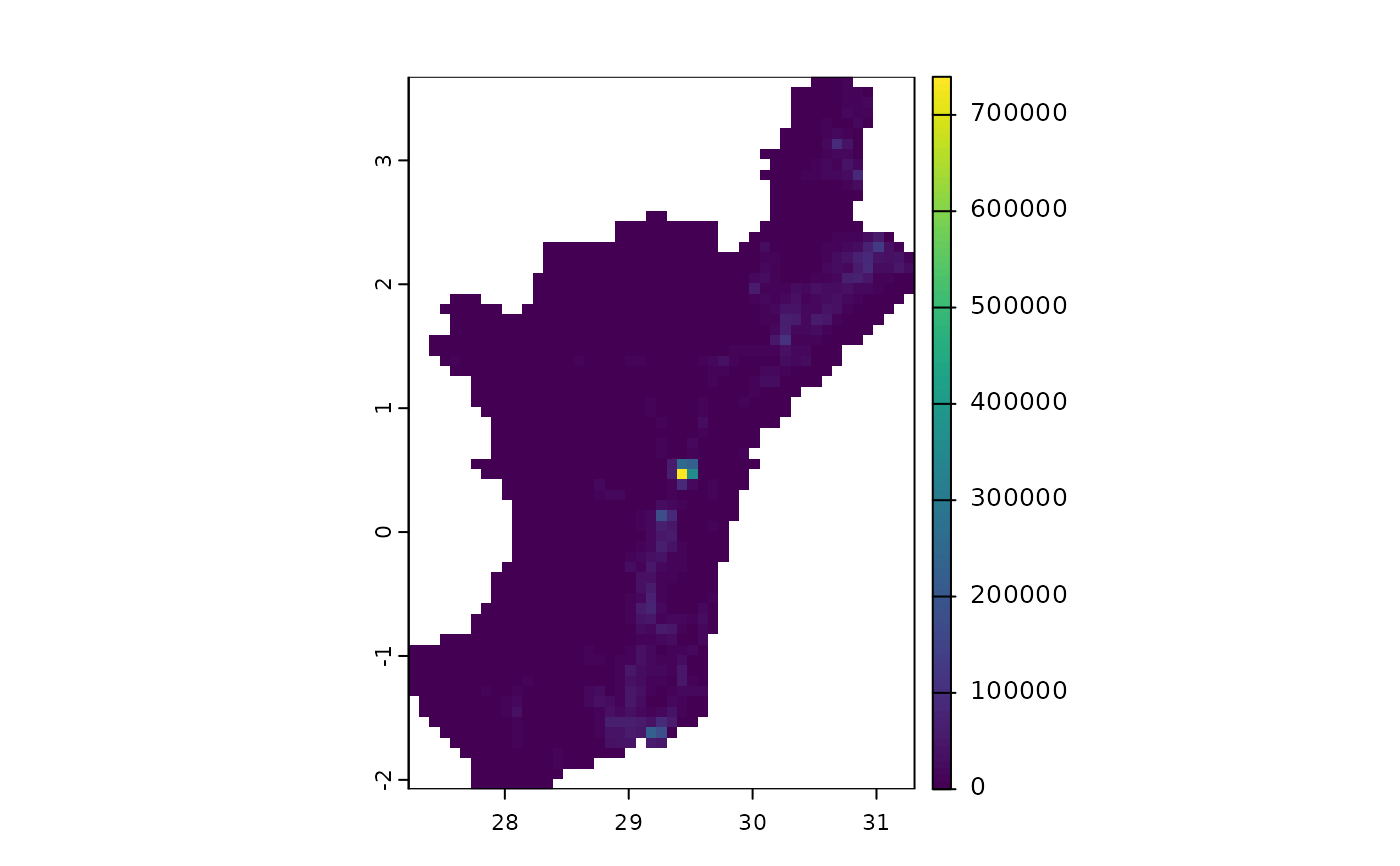 terra::plot(castSeedDataQueensNeighbourhood(layers, initialInfections.fourCities, 1)$Susceptible)
terra::plot(castSeedDataQueensNeighbourhood(layers, initialInfections.fourCities, 1)$Susceptible)